 I am a Research Scientist at the Max Delbrück Center for Molecular Medicine in Berlin, Germany.
My current research focuses on deep machine learning and computer vision.
I obtained my PhD in 2023 at the Humboldt University of Berlin where I was advised by Prof. Dr. Kainmueller.
During this time I developed novel machine learning methods for the instance segmentation and tracking of cells and nuclei in biomedical data and analyzed their theoretical properties.
I received my M.Sc. degree in Computer Science in 2017 at the Dresden University of Technology working with Prof. Dr. Carsten Rother and Dr. Alexander Krull.
We developed an automated framework for Scanning Probe Microscopy based on deep reinforcement learning, published at nature communications physics.
At the same time I worked as a research assistant for the Chair of Compiler Construction.
I completed my B.Sc. degree in Computer Science and Media at the Bauhaus University of Weimar working with Prof. Froehlich on 3d reconstruction using an array of Kinect cameras.
I am a Research Scientist at the Max Delbrück Center for Molecular Medicine in Berlin, Germany.
My current research focuses on deep machine learning and computer vision.
I obtained my PhD in 2023 at the Humboldt University of Berlin where I was advised by Prof. Dr. Kainmueller.
During this time I developed novel machine learning methods for the instance segmentation and tracking of cells and nuclei in biomedical data and analyzed their theoretical properties.
I received my M.Sc. degree in Computer Science in 2017 at the Dresden University of Technology working with Prof. Dr. Carsten Rother and Dr. Alexander Krull.
We developed an automated framework for Scanning Probe Microscopy based on deep reinforcement learning, published at nature communications physics.
At the same time I worked as a research assistant for the Chair of Compiler Construction.
I completed my B.Sc. degree in Computer Science and Media at the Bauhaus University of Weimar working with Prof. Froehlich on 3d reconstruction using an array of Kinect cameras.
Contact
News
[2024] Our paper FISBe: A real-world benchmark dataset for instance segmentation of long-range thin filamentous structures was accepted at CVPR! The dataset is available on zenodo.
[2023] Our paper ACTIS: Improving data efficiency by leveraging semi-supervised Augmentation Consistency Training for Instance Segmentation was published at the BioImage Computing (BIC) workshop at ICCV
[2023] Our preprint Common Limitations of Image Processing Metrics: A Picture Story is available on arXiv
[2023] I successfully defended and published my PhD thesis!
[2023] Our preprint CoNIC Challenge: Pushing the Frontiers of Nuclear Detection, Segmentation, Classification and Counting is available on arXiv (update 2024: now published in Medical Image Analysis)
[2022] Our paper Tracking by Weakly-Supervised Learning and Graph Optimization for Whole-Embryo C. elegans lineages was published at MICCAI
[2022] Our paper Automated reconstruction of whole-embryo cell lineages by learning from sparse annotations was published by Nature Biotechnology
[2022] Our book chapter Mathematical and bioinformatic tools for cell tracking was published as part of the book “Cell Movement in Health and Disease” by Academic Press
[2022] Our preprint PatchPerPixMatch for Automated 3d Search of Neuronal Morphologies in Light Microscopy is available on bioarxiv
[2022] Our paper Panoptic segmentation with highly imbalanced semantic labels was published at ISBIC
[2022] Our method for panoptic segmentation ended up in 2nd place in the CoNIC segmentation and classification challenge
[2022] Our poster “What can go wrong with the tile & stitch?” received a runner-up for best poster award at CBIAS 2021
[2021] Our paper How Shift Equivariance Impacts Metric Learning for Instance Segmentation was published at ICCV
[2020] Our paper An Auxiliary Task for Learning Nuclei Segmentation in 3D Microscopy Images was published at MIDL
[2020] Our paper PatchPerPix for Instance Segmentation was published at ECCV
[2020] Our paper Artificial-intelligence-driven scanning probe microscopy was published by Nature Communications Physics
Datasets
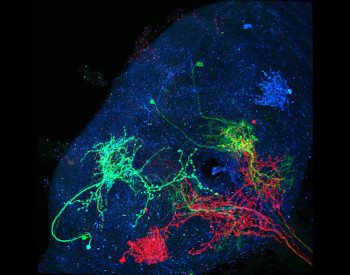 FISBe: A real-world benchmark dataset for instance segmentation of long-range thin filamentous structures
FISBe: A real-world benchmark dataset for instance segmentation of long-range thin filamentous structures
Lisa Mais*, Peter Hirsch*, Claire Managan, Ramya Kandarpa, Josef Lorenz Rumberger, Annika Reinke, Lena Maier-Hein, Gudrun Ihrke, Dagmar Kainmueller, zenodo
[Data]
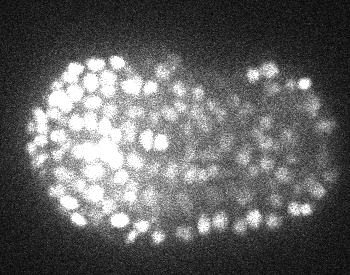 3D+time nuclei tracking dataset of confocal fluorescence microscopy time series of C. elegans embryos
3D+time nuclei tracking dataset of confocal fluorescence microscopy time series of C. elegans embryos
Anthony Santella, Ismar Kovacevic, Zhirong Bao, Peter Hirsch, zenodo
[Data]
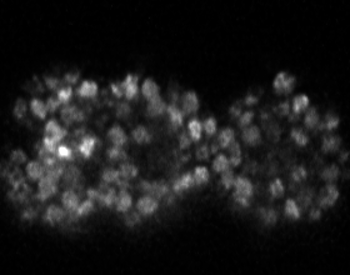 3D+time nuclei tracking dataset of diSPIM lightsheet fluorescence microscopy time series of C. elegans embryos
3D+time nuclei tracking dataset of diSPIM lightsheet fluorescence microscopy time series of C. elegans embryos
Ryan Christensen, Mark W. Moyle, Hari Shroff, Peter Hirsch, zenodo
[Data]
Publications
 FISBe: A real-world benchmark dataset for instance segmentation of long-range thin filamentous structures
Lisa Mais*, Peter Hirsch*, Claire Managan, Ramya Kandarpa, Josef Lorenz Rumberger, Annika Reinke, Lena Maier-Hein, Gudrun Ihrke, Dagmar Kainmueller, CVPR 2024
FISBe: A real-world benchmark dataset for instance segmentation of long-range thin filamentous structures
Lisa Mais*, Peter Hirsch*, Claire Managan, Ramya Kandarpa, Josef Lorenz Rumberger, Annika Reinke, Lena Maier-Hein, Gudrun Ihrke, Dagmar Kainmueller, CVPR 2024
[PDF] [Project Page/Code]
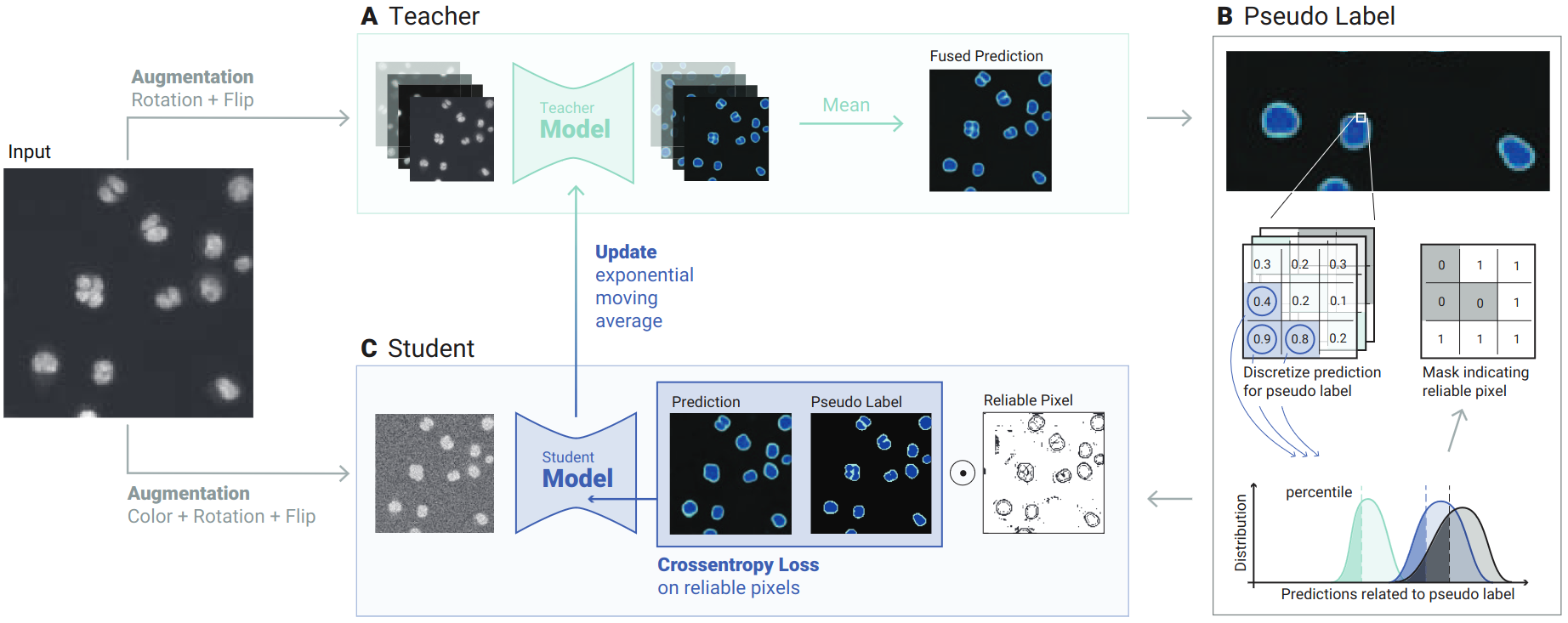 ACTIS: Improving data efficiency by leveraging semi-supervised Augmentation Consistency Training for Instance Segmentation
Josef Lorenz Rumberger, Jannik Franzen, Peter Hirsch, Jan-Philipp Albrecht, Dagmar Kainmueller, ICCV BIC Workshop 2023
ACTIS: Improving data efficiency by leveraging semi-supervised Augmentation Consistency Training for Instance Segmentation
Josef Lorenz Rumberger, Jannik Franzen, Peter Hirsch, Jan-Philipp Albrecht, Dagmar Kainmueller, ICCV BIC Workshop 2023
[PDF] [Project Page/Code]
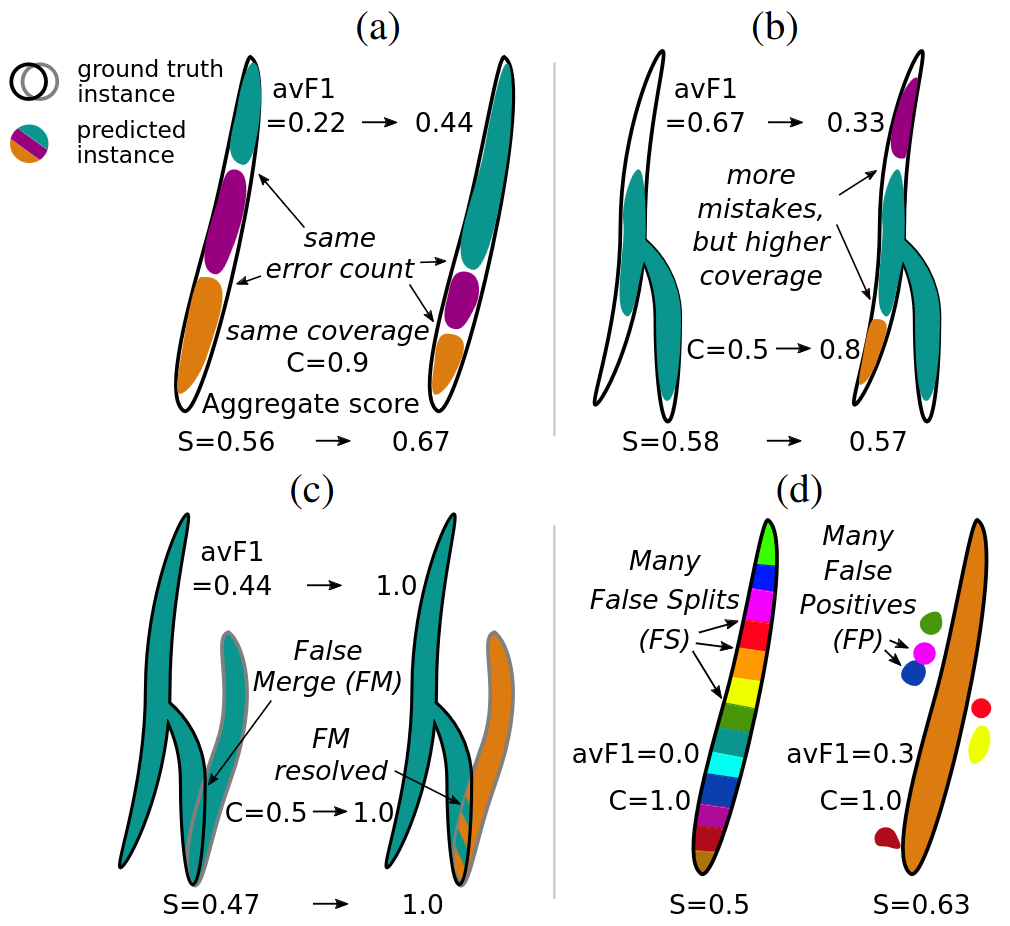
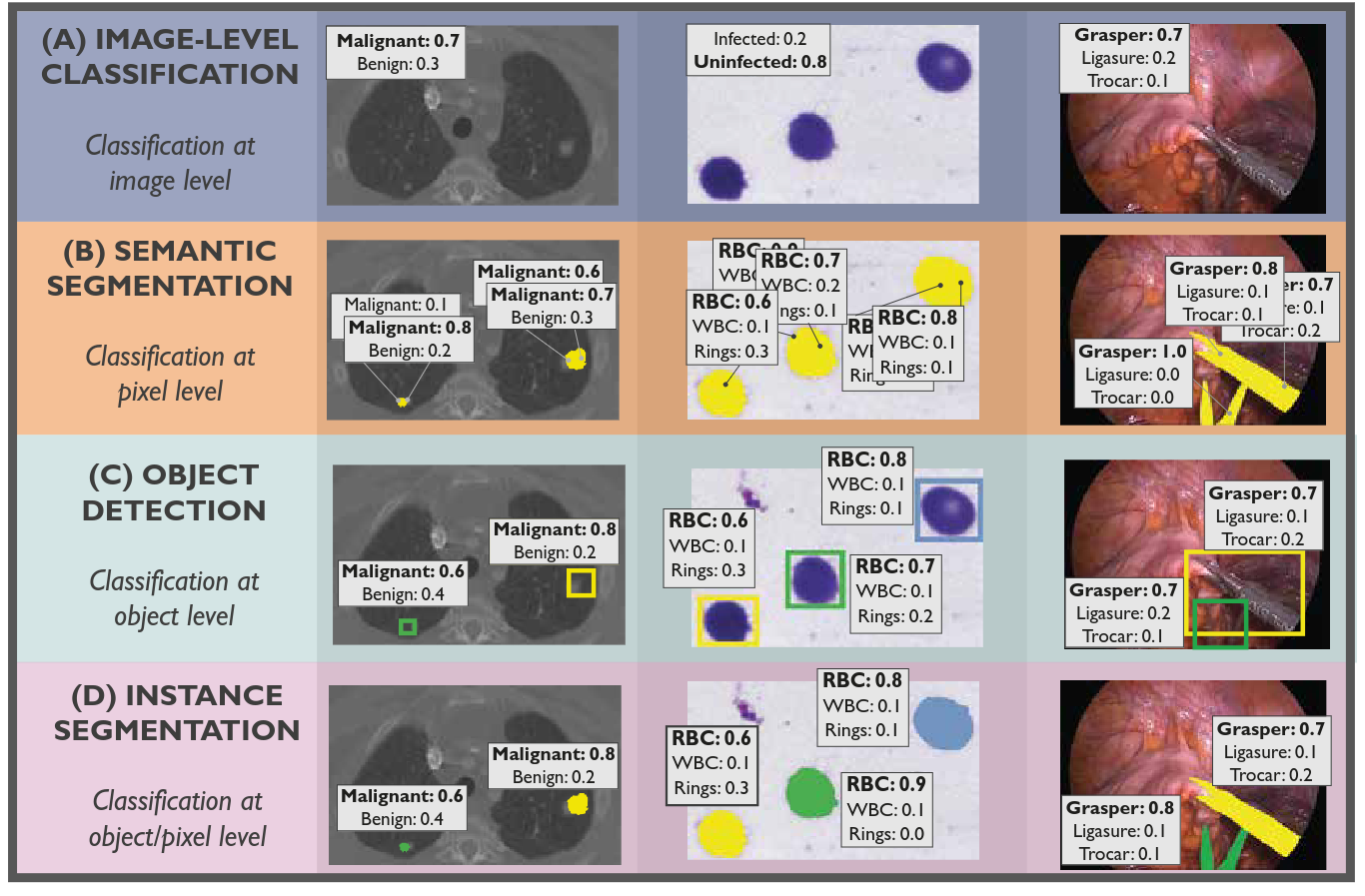 Common Limitations of Image Processing Metrics: A Picture Story
Common Limitations of Image Processing Metrics: A Picture Story
Annika Reinke, Minu D. Tizabi, …, Peter Hirsch, …, Lena Maier-Hein, arXiv
[PDF]
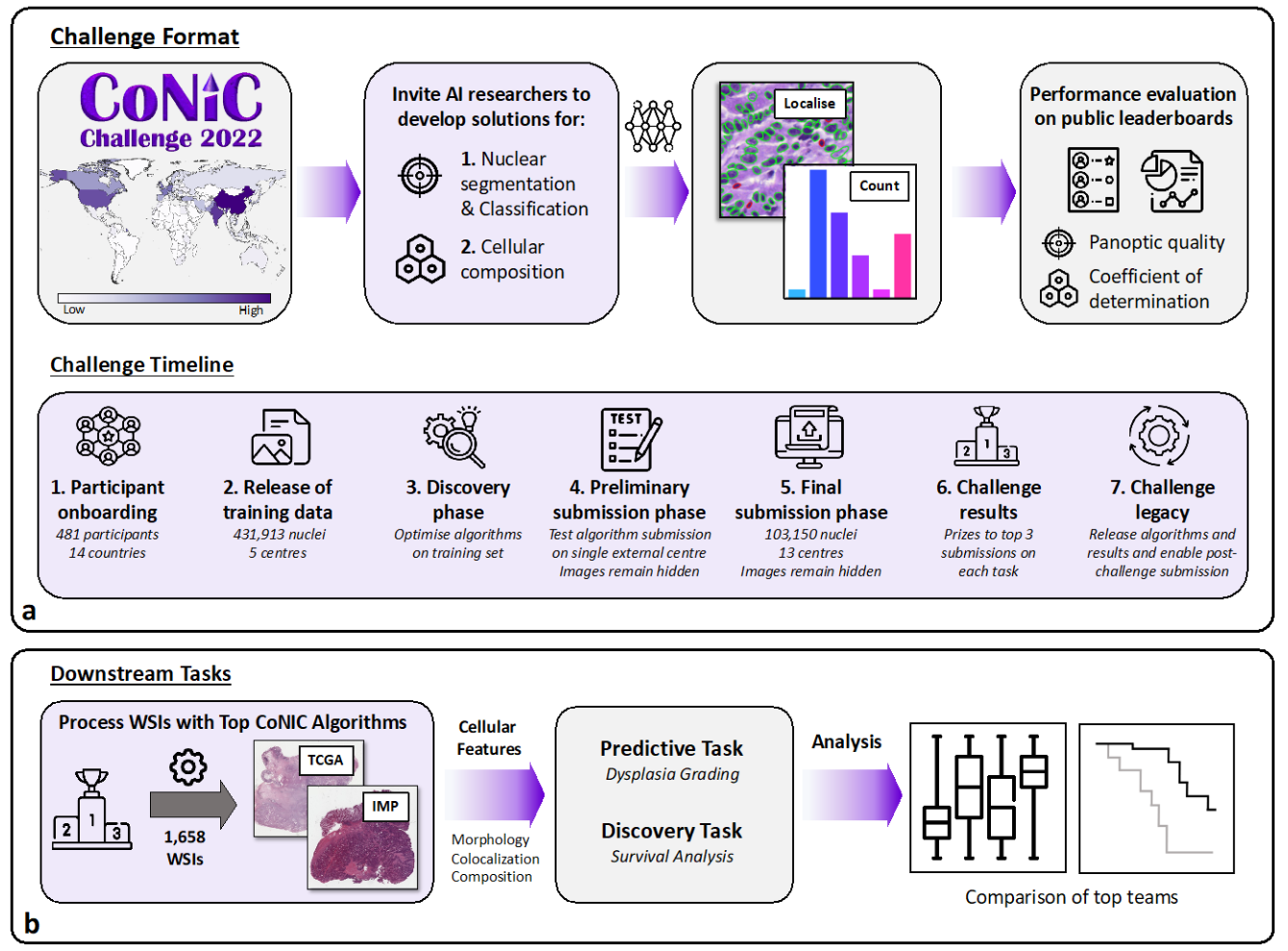 CoNIC Challenge: Pushing the Frontiers of Nuclear Detection, Segmentation, Classification and Counting
CoNIC Challenge: Pushing the Frontiers of Nuclear Detection, Segmentation, Classification and Counting
Simon Graham, Quoc Dang Vu, Mostafa Jahanifar, …, Peter Hirsch, …, David Snead, Shan E. Ahmed Raza, Fayyaz Minhas, Nasir M. Rajpoot, arXiv
[PDF]
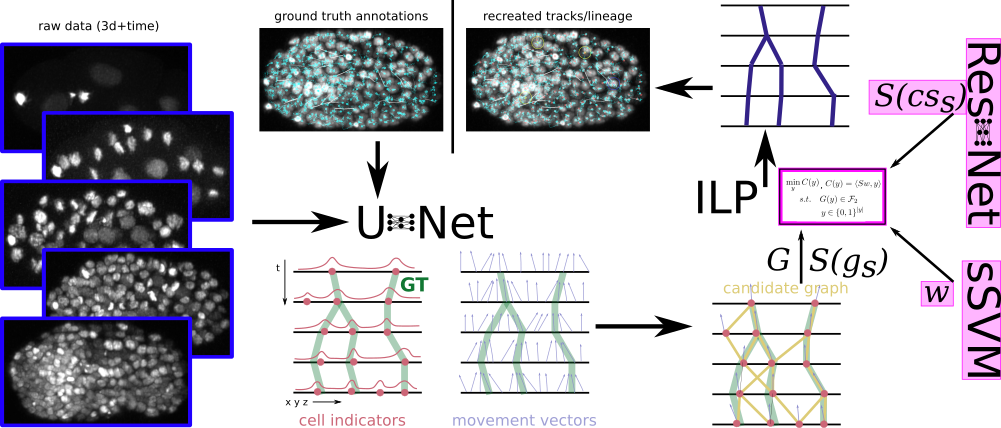 Tracking by Weakly-Supervised Learning and Graph Optimization for Whole-Embryo C. Elegans lineages
Tracking by Weakly-Supervised Learning and Graph Optimization for Whole-Embryo C. Elegans lineages
Peter Hirsch, Caroline Malin-Mayor, Anthony Santella, Stephan Preibisch, Dagmar Kainmüller, and Jan Funke, MICCAI 2022
[PDF] [Project Page/Code]
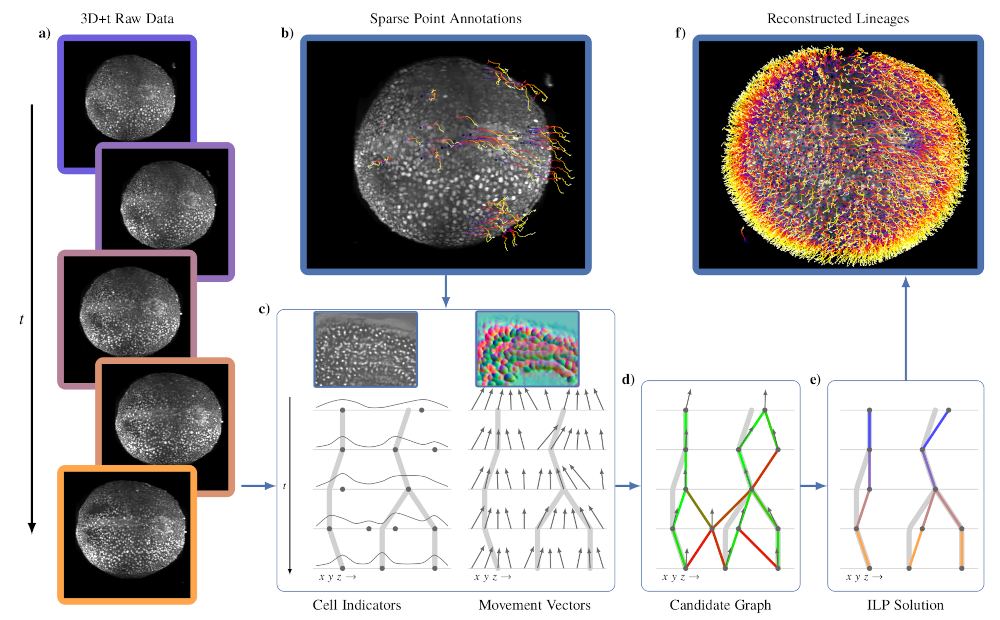 Automated reconstruction of whole-embryo cell lineages by learning from sparse annotations
Automated reconstruction of whole-embryo cell lineages by learning from sparse annotations
Caroline Malin-Mayor, Peter Hirsch, Leo Guignard, Katie McDole, Yinan Wan, William C. Lemon, Dagmar Kainmueller, Philipp J. Keller, Stephan Preibisch, Jan Funke, Nature Biotechnology 2022
[PDF] [Project Page/Code]
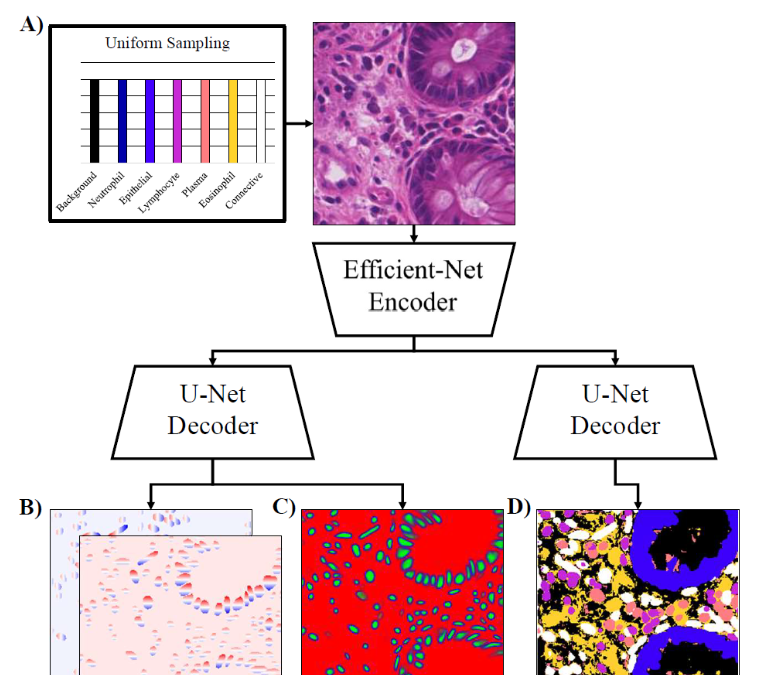 Panoptic segmentation with highly imbalanced semantic labels
Panoptic segmentation with highly imbalanced semantic labels
Josef Lorenz Rumberger*, Elias Baumann*, Peter Hirsch*, Andrew Janowczyk, Inti Zlobec, and Dagmar Kainmueller, ISBIC 2022 (2nd place in the CoNIC segmentation and classification challenge)
[PDF]
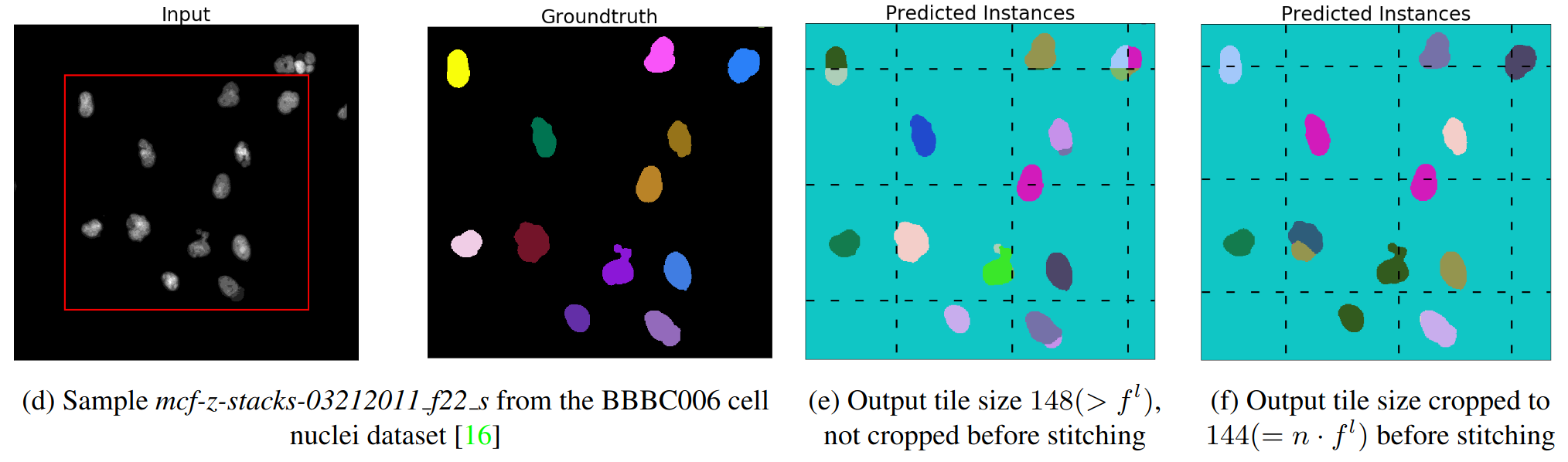 How Shift Equivariance Impacts Metric Learning for Instance Segmentation
How Shift Equivariance Impacts Metric Learning for Instance Segmentation
Josef Lorenz Rumberger*, Xiaoyan Yu*, Peter Hirsch*, Melanie Dohmen*, Vanessa Emanuela Guarino*, Ashkan Mokarian, Lisa Mais, Jan Funke, Dagmar Kainmüller, ICCV 2021 (also presented at Crick BioImage Analysis Symposium 2021 as “What can go wrong with the tile & stitch?” (runner-up for best poster))
[PDF] [Project Page]
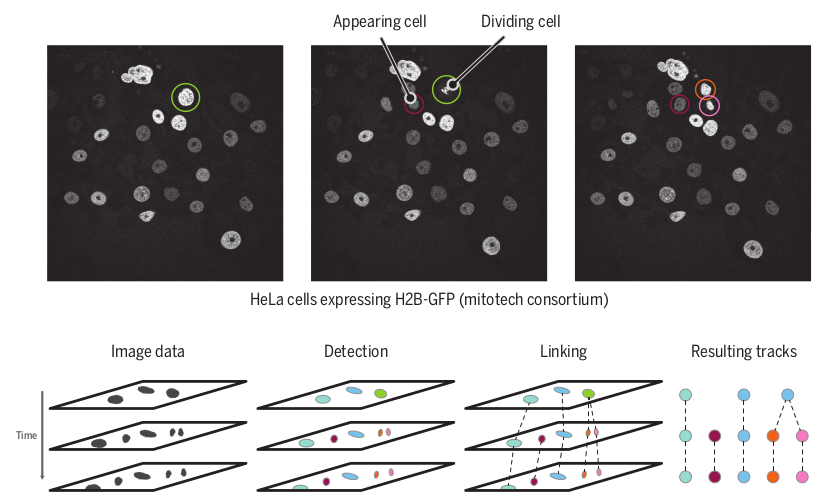 Chapter 20 - Mathematical and bioinformatic tools for cell tracking
Chapter 20 - Mathematical and bioinformatic tools for cell tracking
Peter Hirsch, Leo Epstein, and Léo Guignard, Cell Movement in Health and Disease, Academic Press 2022
[PDF]
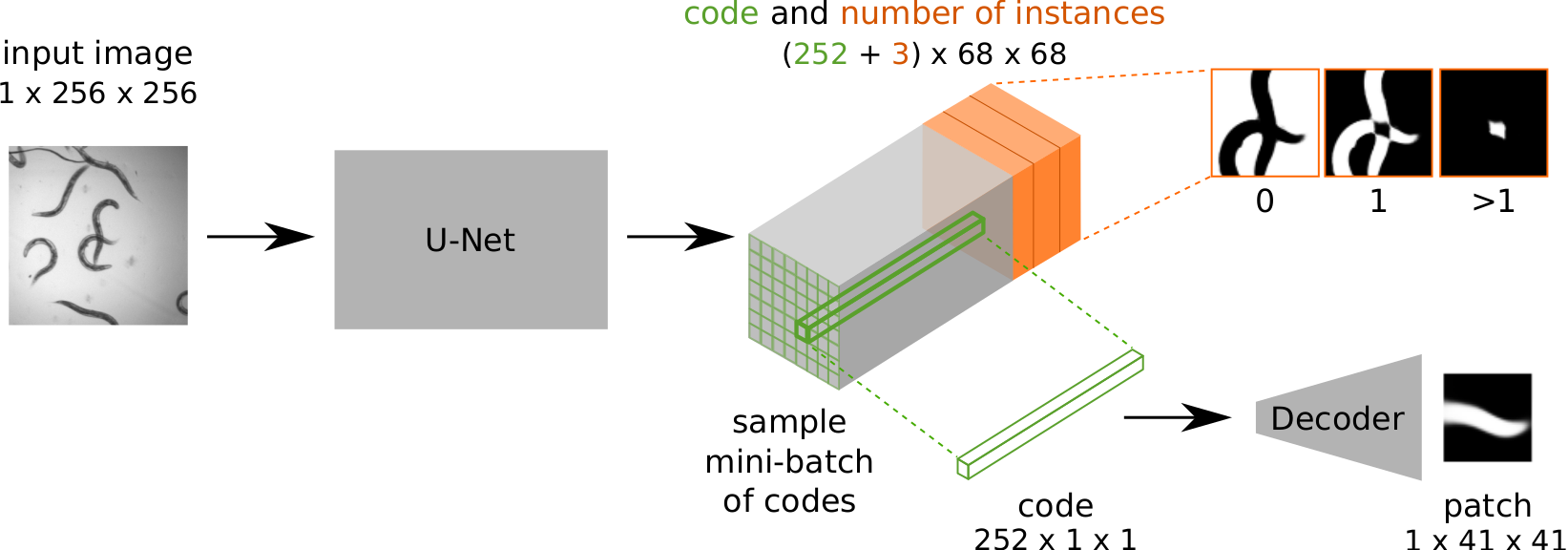 PatchPerPix for Instance Segmentation
PatchPerPix for Instance Segmentation
Peter Hirsch*, Lisa Mais*, Dagmar Kainmueller, ECCV 2020
[PDF] [Project Page/Code]
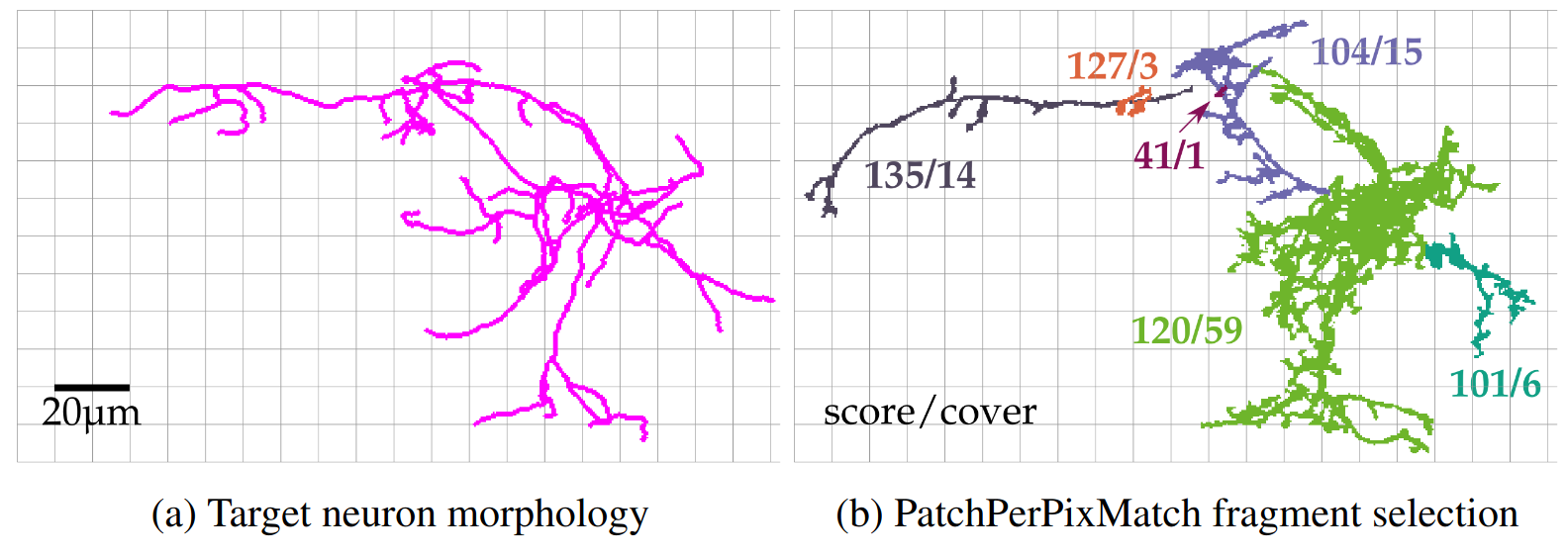 PatchPerPixMatch for Automated 3d Search of Neuronal Morphologies in Light Microscopy
PatchPerPixMatch for Automated 3d Search of Neuronal Morphologies in Light Microscopy
Lisa Mais, Peter Hirsch, Claire Managan, Kaiyu Wang, Konrad Rokicki, Robert R. Svirskas, Barry J. Dickson, Wyatt Korff, Gerald M. Rubin, Gudrun Ihrke, Geoffrey W. Meissner, and Dagmar Kainmüller, bioraxiv/preprint
[PDF]
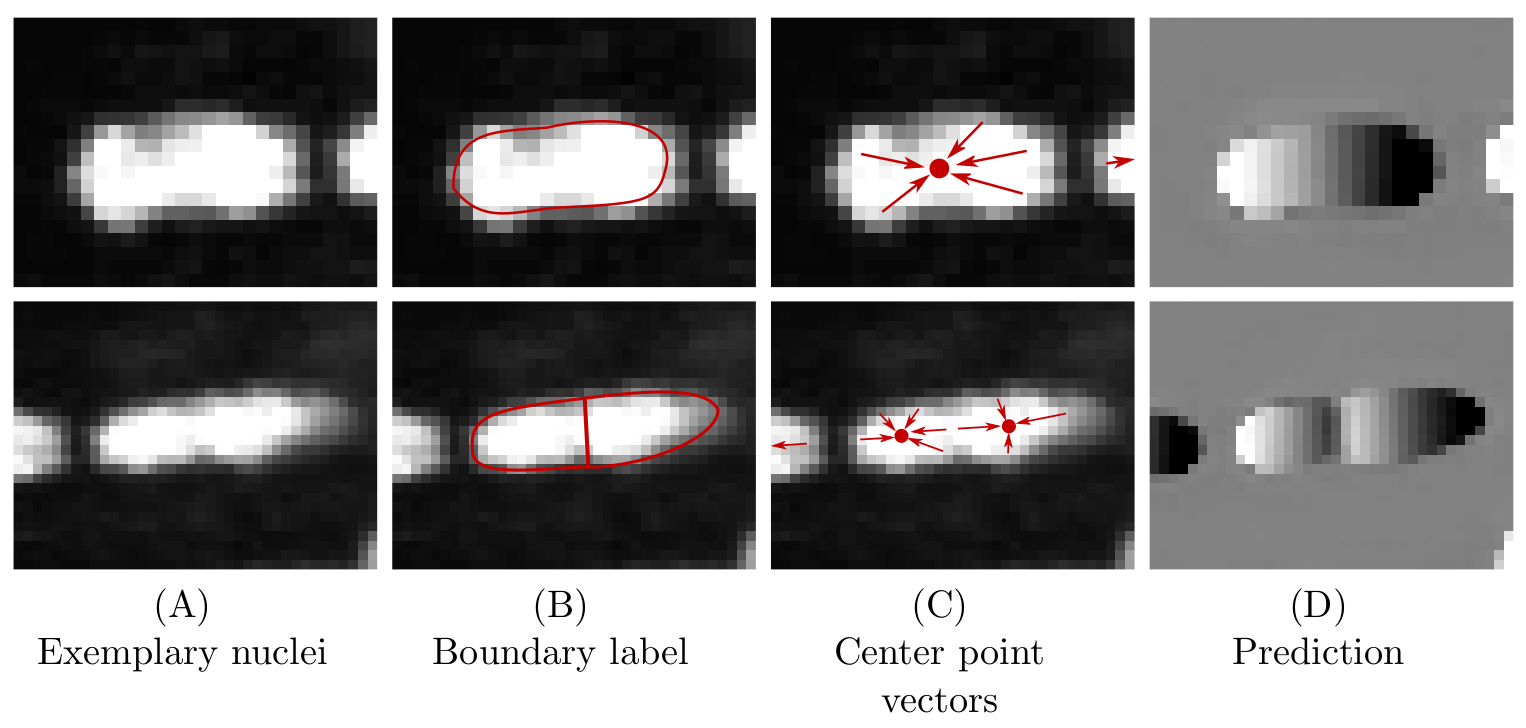 An Auxiliary Task for Learning Nuclei Segmentation in 3D Microscopy Images
An Auxiliary Task for Learning Nuclei Segmentation in 3D Microscopy Images
Peter Hirsch, Dagmar Kainmüller, MIDL 2020
[PDF] [Project Page/Code]
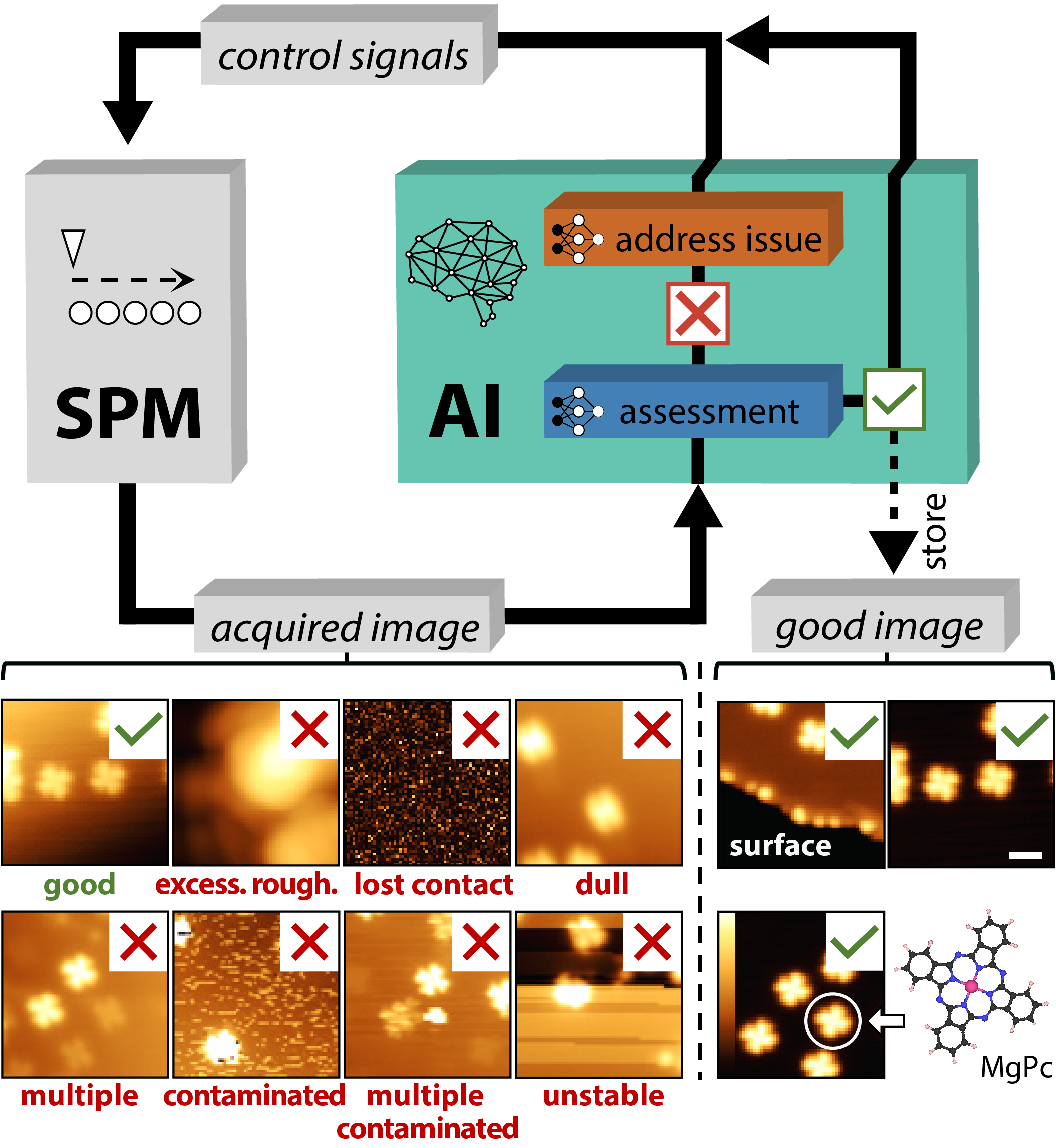 Artificial-intelligence-driven scanning probe microscopy
Artificial-intelligence-driven scanning probe microscopy
Alexander Krull*, Peter Hirsch*, Carsten Rother, Agustin Schiffrin, and Cornelius Krull, Nature Communications Physics 2020
[PDF] [Project Page/Code]